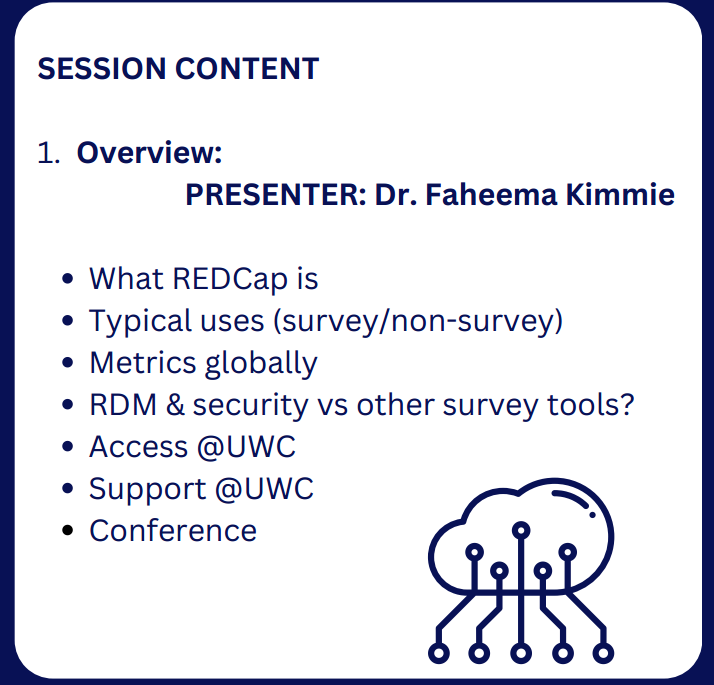
The eResearch Office at UWC is a division under the Deputy Vice-Chanellor Research and Innovation that promotes the use of information technology to enable fast and high impact research. Part of its mission is to drive and coordinate advanced uses of data-driven approaches to multi-, inter- and trans-disciplinary research in a dynamic environment for skills development and academic growth. A postdoctoral fellowship position is available for an outstanding candidate to undertake cutting-edge research in the development of data technology for sustainable food systems. The fellow will be awarded a tax-free fellowship for up to 2 years with opportunities for academic and personal development.
Prerequisites:
The prospective candidate should be a holder of a doctoral degree obtained within the past five years in the field related to Natural, Computer, Mathematical or Agricultural Sciences. The applicant should have interest and/or experience in modelling data of biological systems, be familiar with the concepts of sustainability within food systems and able to work without supervision, as well as in a team.
The successful candidate will be involved in the following roles:
– Conducting research in the area of data science applied to food systems
– Develop original research and publication
– Involvement in sourcing research grants
– Science engagement
– May be requested to assist with various activities that support the eResearch agenda for the University.
– Mentor and support postgraduate students.
Interested candidates can send their CV and motivation letter to Dr Frederic Isingizwe at fisingizwe@uwc.ac.za or to Prof Clement Nyirenda at cnyirenda@uwc.ac.za.
The position remains open until filled.